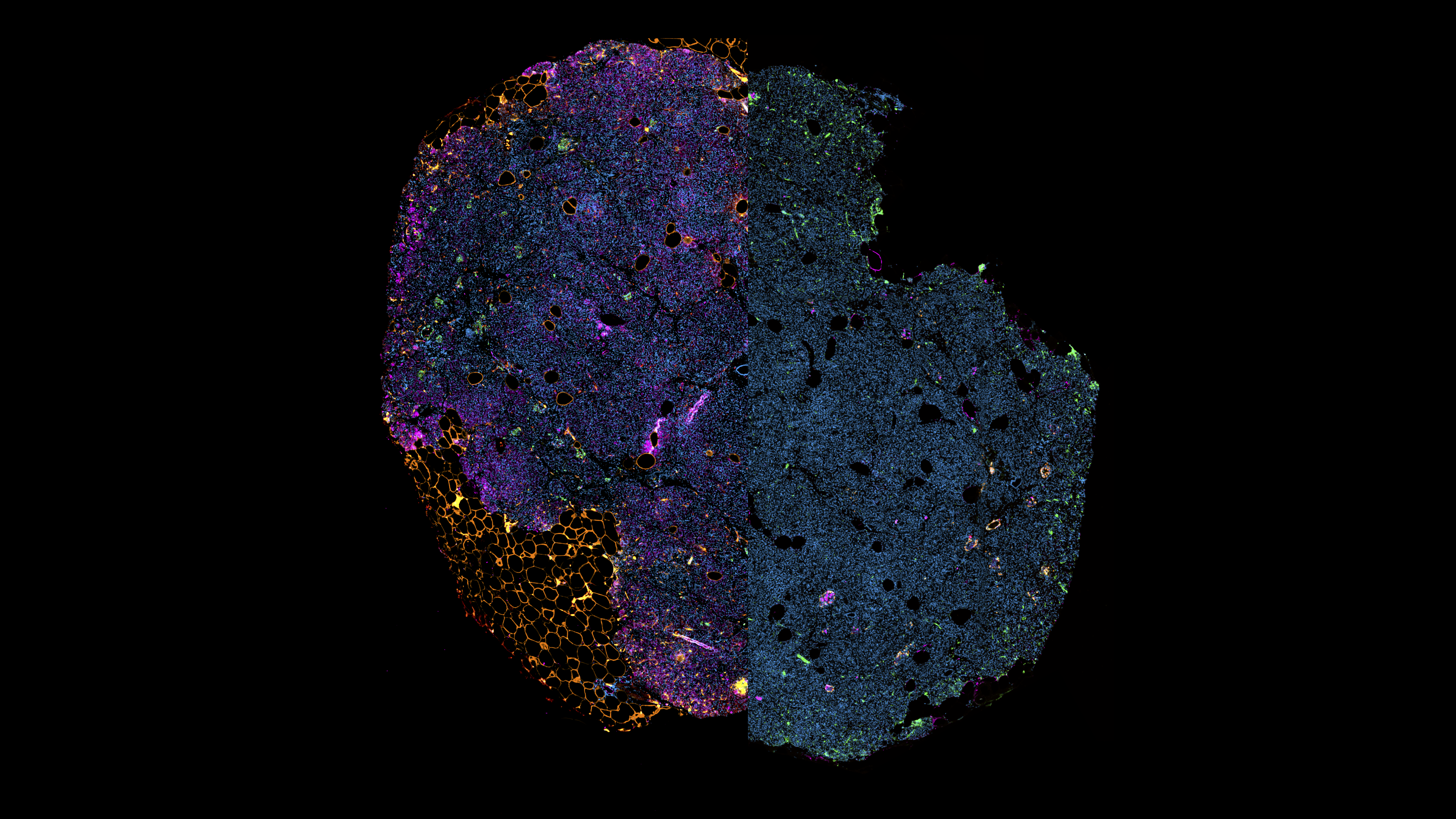
High-Resolution Multiplex Dataset of the Human Pancreas in Type 2 Diabetes
Paper | Data (see "Data Access" section for password) | Cite
Introduction
Type 2 diabetes affects approximately 500 million people worldwide, yet our ability to detect the disease through tissue analysis remains limited. Traditional histopathological approaches struggle to identify the subtle morphological changes associated with impaired insulin secretion and β-cell failure. The DIADEM (type 2 DIAbetes DEtection) dataset was created to address this gap by providing high-resolution pancreatic tissue images that capture alterations invisible to conventional analysis. This comprehensive collection includes whole-slide images from living donors with multiple chromogenic IHC and multiplex immunofluorescence (mIF) stainings, paired with clinical data to support both machine learning development and clinical research.
The dataset enables researchers to explore computationally discovered biomarkers linked to type 2 diabetes status and other clinical variables. Deep learning models trained on this data have revealed that predictive signals emerge from islet α- and δ-cells, neuronal axons, adipocyte cluster size, islet-adipocyte proximity, and islet dimensions. By making these learned relationships interpretable through explainable AI techniques, the DIADEM dataset offers a foundation for investigating diagnostic and therapeutic targets while refining hypotheses about pancreatic tissue alterations in type 2 diabetes.
Data Overview
The dataset consists of three parts: the IHC data, the mIF data and the clinical patient data.
The clinical patient data is pseudonymised with a patient identifier in the format of PXXX (e.g. P100).
The mIF data contains stainings visualizing cells within the pancreatic islets as well as other cell types related to type 2 diabetes hypotheses.
It consists of two staining sets (see below) and two samples per patient (i.e. P100_S1 and P100_S2).
However, some czi files contain both samples in one file, then the file is named PXXX_S1_S2.czi.
The IHC data includes the same six specific stainings as the mIF data, but only one sample per patient and staining.
All tissues for one patients are consecutive cut and exhibit comparable morphological features between each other.
For more information about the cohort and the acquisition of the samples, we refer to our publication.
Multiplex Immunofluorescence (mIF) Staining Sets
The dataset includes two distinct multiplex immunofluorescence staining sets optimized for comprehensive tissue characterization and profiling of the pancreatic islets.
mIF Staining Set 1
- CH1 - Brightfield
- CH2 (AF647) - Tubulin-b3
- CH3 (AF555) - Glucagon
- CH4 - DAPI (Nuclear counterstain)
- CH5 (AF750) - Somatostatin (SST)

mIF Staining Set 2
- CH1 - Brightfield
- CH2 (AF555) - Perilipin-1
- CH3 (AF488) - Insulin
- CH4 - DAPI (Nuclear counterstain)
- CH5 (AF750) - CD31
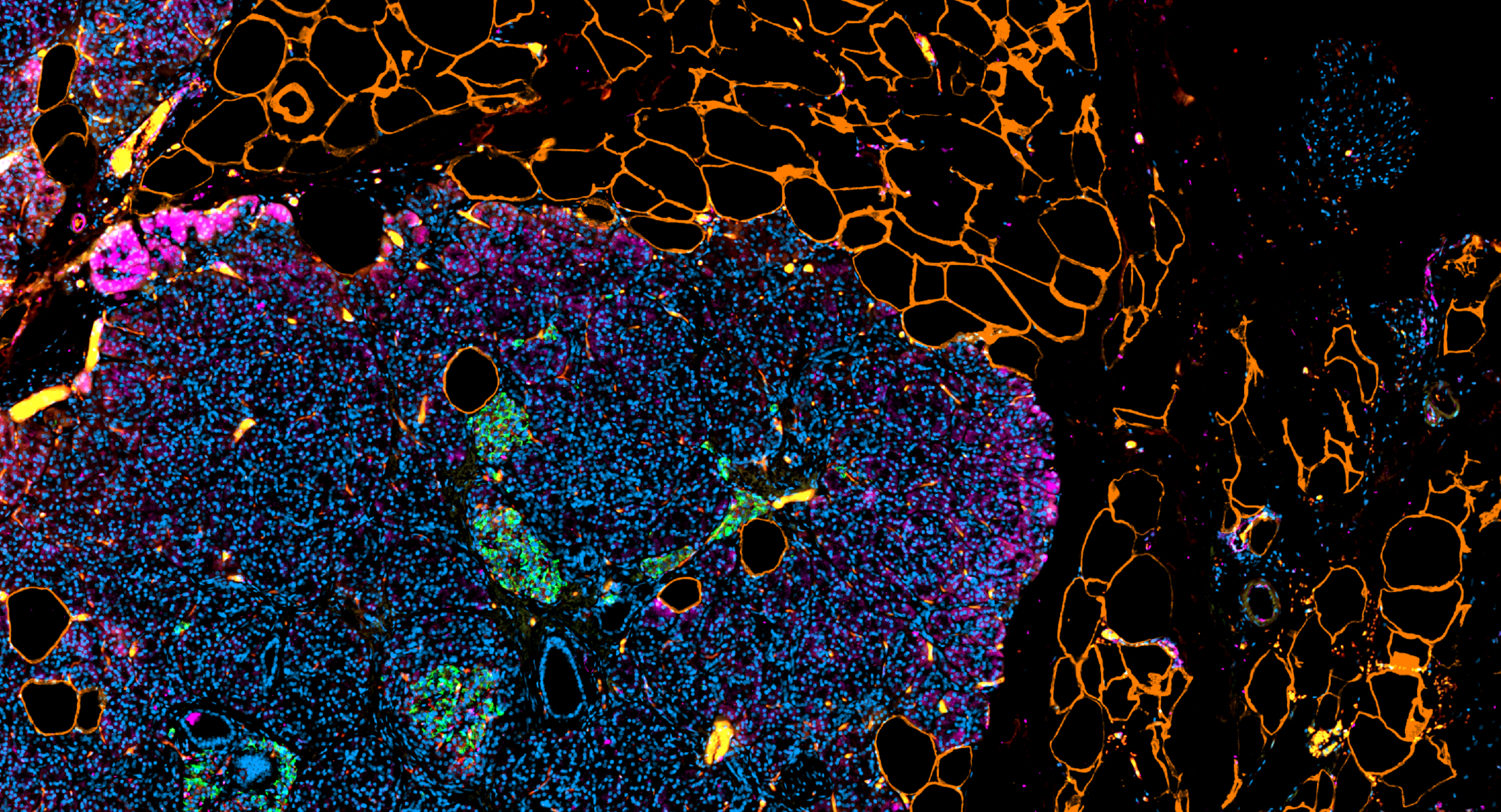
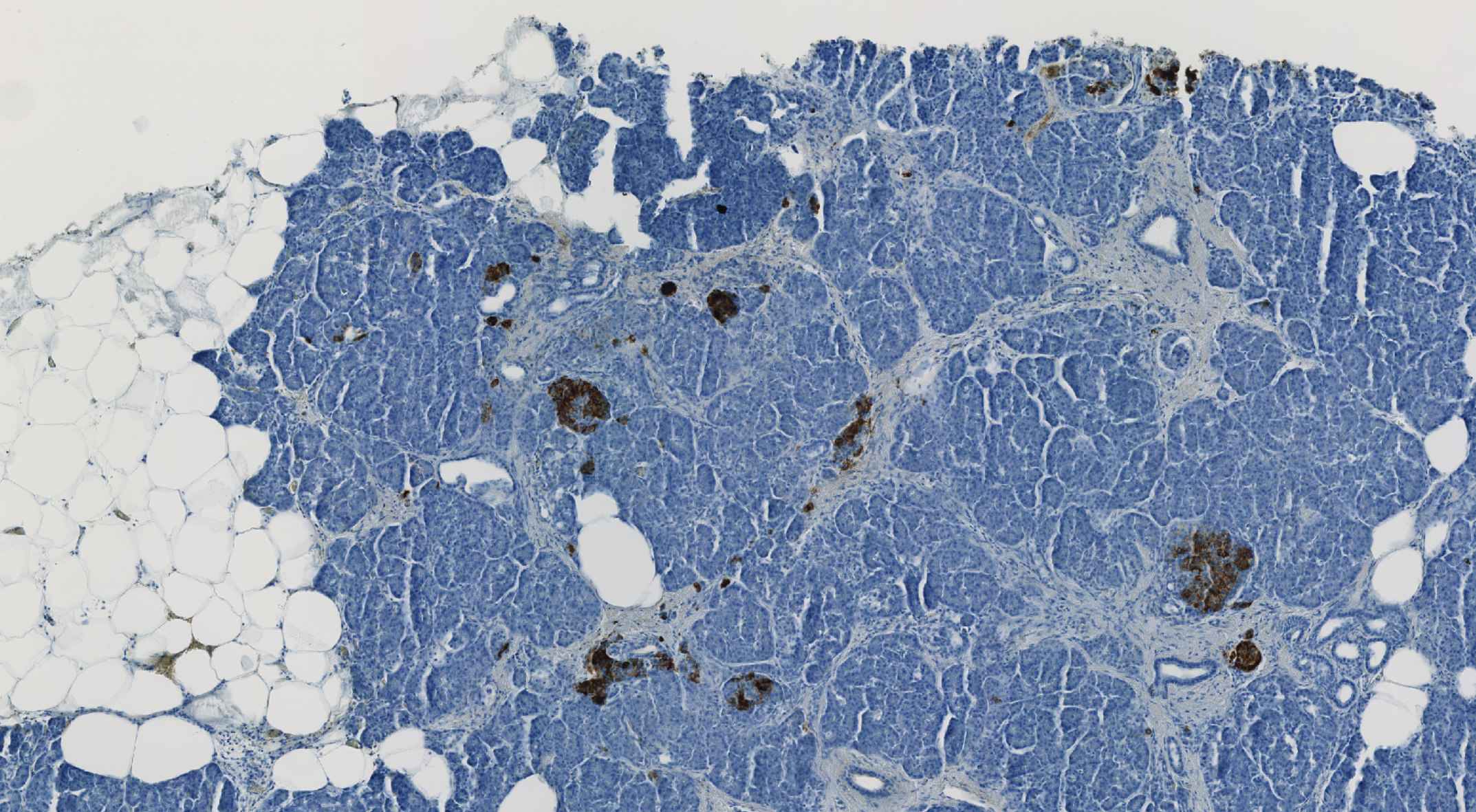
Immunohistochemistry (IHC) Stainings
IHC slides using the same six stainings as the mIF staining sets provide complementary single-marker analysis for validation and cross-method comparison.
IHC Staining Set
- CD31
- Glucagon
- Insulin
- Perilipin-1
- Somatostatin (SST)
- Tubulin-b3







Data Access
The DIADEM dataset is publicly available for research and educational purposes.
Download the complete dataset or single images using the link below. To access the data please use this password:

| Data Type | Formats | Size | Access |
|---|---|---|---|
| Complete Dataset | CZI, NDPI, XLSX | 735 GB | Download |
Citation & Data Usage Policy
Citation
Klein, L., Ziegler, S., Gerst, F., Morgenroth, Y. et al. Explainable AI-based analysis of human pancreas sections identifies traits of type 2 diabetes. Nat Commun (2025).
Data Usage Policy
The DIADEM dataset is released under the Commons Attribution–NonCommercial 4.0 (CC BY-NC 4.0) license for research and educational purposes. Users must cite the dataset appropriately in all publications and presentations. The data has been de-identified in accordance with applicable regulations. By downloading or using this dataset, you agree to abide by these terms and handle the data responsibly.
Acknowledgments
The DIADEM dataset was developed through the collaborative efforts of multiple institutions and research teams. We gratefully acknowledge the following contributions:
- We thank all the participants of the "LIDOPACO" programs in Tübingen and Dresden. The studies were supported by the German Center for Diabetes Research (Deutsches Zentrum für Diabetesforschung, DZD). The DZD is funded by the German Federal Ministry for Education and Research and the states where its partner institutions are located (01GI0925).
- The authors acknowledge the project specific financial support of the Helmholtz Association (project DIADEM, ZT-1-PF-5 139).
- This project has received funding from the European Union's Horizon Europe research and innovation program under grant agreement No 1010954433 (Intercept-T2D). Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union. Neither the European Union nor the granting authority can be held responsible for them.
- This work was funded by Helmholtz Imaging (HI), a platform of the Helmholtz Incubator on Information and Data Science.
- Birkenfeld A. was supported by the German Federal Ministry for Education and Research (01GI0925) via the German Center for Diabetes Research (DZD eV); Ministry of Science, Research and the Arts Baden-Württemberg; and Helmholtz Munich.
- This work was supported by the Light Microscopy Facility, a Core Facility of the CMCB Technology Platform at TU Dresden.
Contact Information
For questions or collaboration opportunities, please contact:
Email: robert.wagner@uni-duesseldorf.de, michele.solimena@tu-dresden.de, or lukas.klein@epfl.ch.